Estimate distribution of true positives given sampling resuts
Source:R/finite_pop_sampling.R
truepos_given_sample.RdEstimate distribution of true positives given sampling resuts
truepos_given_sample(samplepos, n, N, replicates = 1000) # S3 method for truepos summary(object, alpha = 0.1, ...)
Arguments
| samplepos | Number of positives observed in sample |
|---|---|
| n | Sample size |
| N | Population size |
| replicates | Number of replicates per tested true pos number |
| object | Sample counts to summarise |
| alpha | The confidence interval is (1-alpha)*100% (i.e. alpha=0.1 => 90% CI) |
| ... | Additional arguments (currently ignored) |
Value
a vector containing population true positive counts that could have
generated the observed number of sample positives. It has class
truepos.
Details
The idea is to generate random realisations for all possible numbers of true positives, choose only those cases that resulted in the observed number of sample positives, and then use that empirical distribution of simulated true positives to estimate the most likely value of the (unknown) number of true positives.
NB what we are doing here effectively is to estimate the unknown parameter,
m, of the Hypergeometric distribution, i.e.
the number of white balls in the urn.
See also
Other population-sampling: prop.ci,
required.sample.size,
sample_finite_population
Examples
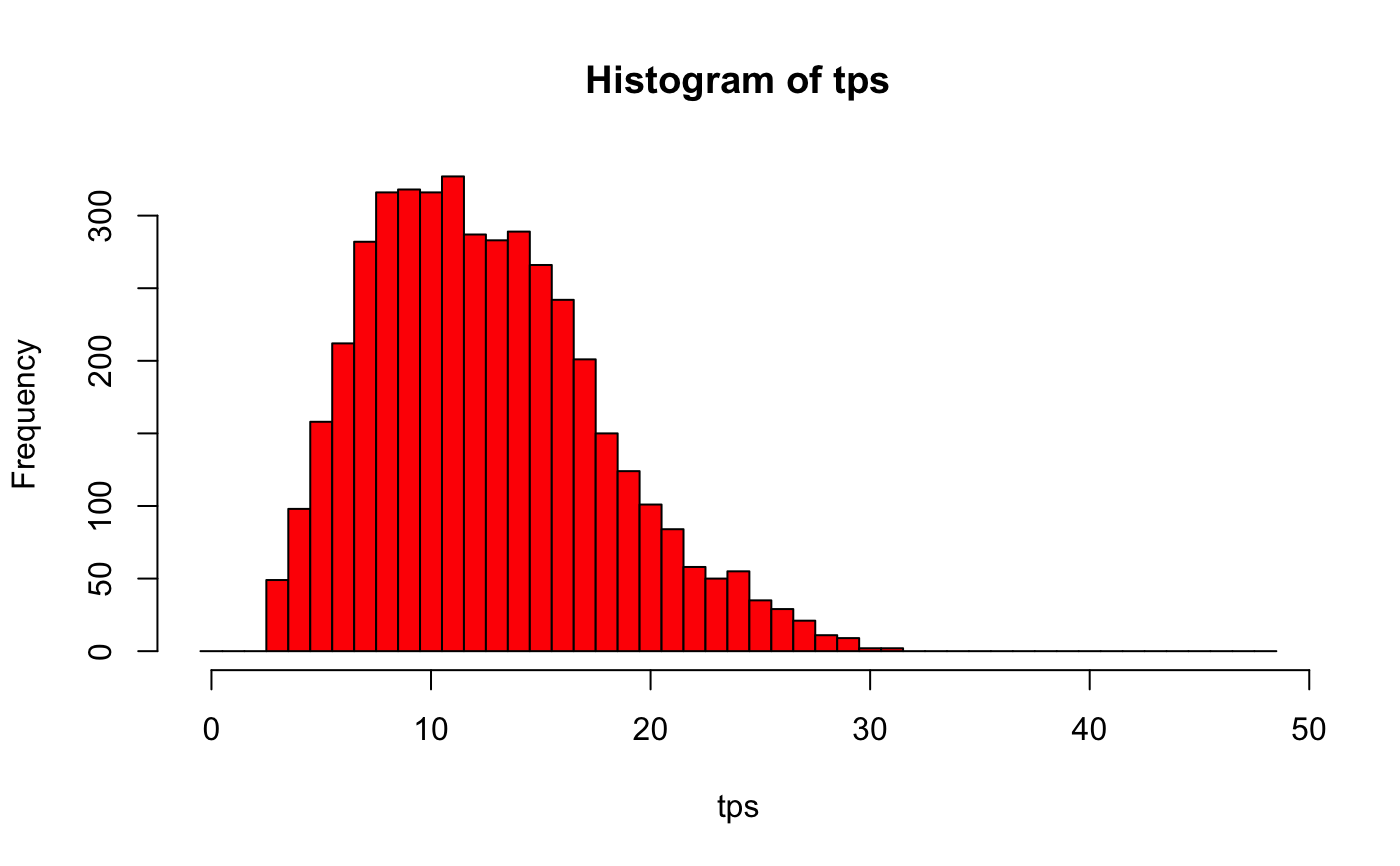
# Imagine we have sampled 10 profiles from a tract of 48 and found 2 LHNs tps=truepos_given_sample(samplepos = 2, n=10, N=48) hist(tps, breaks=0:49-.5, col='red')# the mode should be the Maximum Likelihood Estimate # (if enough replicates were used) summary(tps)#> 5% Median Mode 95% #> 5 12 11 22#> 2.5% Median Mode 97.5% #> 4 12 11 24